Basic Usage¶
- The two required input elements to run diffusion using DiffuPy are:
A network/graph. (see Network-Input Formatting below)
A dataset of scores. (see Scores-Input Formatting below)
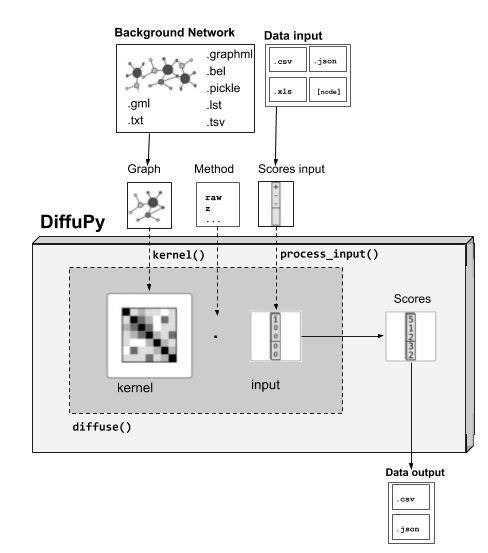
For its usability, you can either:
Use the Command Line Interface (see cli).
Use pythonically the functions provided in diffupy.diffuse:
from diffupy.diffuse import run_diffusion
# DATA INPUT and GRAPH as PATHs -> returned as *PandasDataFrame*
diffusion_scores = run_diffusion(~/data/input_scores.csv, ~/data/network.csv).as_pd_dataframe()
# DATA INPUT and GRAPH as Python OBJECTS -> exported *as_csv*
diffusion_scores = run_diffusion(input_scores, network).as_csv('~/output/diffusion_results.csv')
Methods¶
The diffusion method by default is z, which statistical normalization has previously shown to outperform. Further parameters to adapt the propagation procedure are also provided, such as choosing from the available diffusion methods or providing a custom method function. See diffusion Methods and/or Method modularity.
diffusion_scores_select_method = run_diffusion(input_scores, network, method = 'raw')
from networkx import page_rank # Custom method function
diffusion_scores_custom_method = run_diffusion(input_scores, network, method = page_rank)
You can also provide your own kernel method or select among the ones provided in the kernels.py function which you can provide as a kernel_method argument. By default regularised_laplacian_kernel is used.
from diffupath.kernels import p_step_kernel # Custom kernel calculation function
diffusion_scores_custom_kernel_method = run(input_scores, method = 'raw', kernel_method = p_step_kernel)
So method stands for the diffusion process method, and kernel_method for the kernel calculation method.
Formatting¶
Before running diffusion algorithms on your network using DiffuPy, take into account the graph and input data/scores formats. You can find specified here samples of supported input scores and networks.
Input format¶
The input is preprocessed and further mapped before the diffusion. See input mapping or see process_input docs for further details. Here we outline the input formats covered for its preprocessing.
Scores¶
You can submit your dataset in any of the following formats:
CSV (.csv)
TSV (.tsv)
pandas.DataFrame
List
Dictionary
(check Input dataset examples)
So you can either provide a path to a .csv or .tsv file:
from diffupy.diffuse import run_diffusion
diffusion_scores_from_file = run_diffusion('~/data/diffusion_scores.csv', network)
or Pythonically as a data structure as the input_scores parameter:
data = {'Node': ['A', 'B',...],
'Node Type': ['Metabolite', 'Gene',...],
....
}
df = pd.DataFrame (data, columns = ['Node','Node Type',...])
diffusion_scores_from_dict = run_diffusion(df, network)
Please ensure that the dataset minimally has a column ‘Node’ containing node IDs. You can also optionally add the following columns to your dataset:
NodeType
LogFC *
p-value
- *
Log2 fold change
Networks¶
If you would like to submit your own networks, please ensure they are in one of the following formats:
BEL (.bel)
CSV (.csv)
GML (.gml or .xml)
GraphML (.graphml or .xml)
Pickle (.pickle). BELGraph object from PyBEL 0.13.2
TSV (.tsv)
TXT (.txt)
Minimally, please ensure each of the following columns are included in the network file you submit:
Source
Target
Optionally, you can choose to add a third column, “Relation” in your network (as in the example below). If the relation between the Source and Target nodes is omitted, and/or if the directionality is ambiguous, either node can be assigned as the Source or Target.
Kernel¶
If you dispose of a precalculated kernel, you can provide directly the kernel object without needing to also provide a graph object. As mentioned above, if you wish to use your kernel method function you can provide it as kernel_method argument on the previous described function.
Input dataset examples¶
DiffuPath accepts several input formats which can be codified in different ways. See the diffusion scores summary for more details on how the labels input are treated according each available method.
1. You can provide a dataset with a column ‘Node’ containing node IDs.
Node |
|---|
A |
B |
C |
D |
from diffupy.diffuse import run_diffusion
diffusion_scores = run_diffusion(dataframe_nodes, network)
Also as a list of nodes:
['A', 'B', 'C', 'D']
diffusion_scores = run_diffusion(['A', 'B', 'C', 'D'], network)
2. You can also provide a dataset with a column ‘Node’ containing node IDs as well as a column ‘NodeType’, indicating the entity type of the node to run diffusion by entity type.
Node |
NodeType |
|---|---|
A |
Gene |
B |
Gene |
C |
Metabolite |
D |
Gene |
Also as a dictionary of type:list of nodes :
{'Gene': ['A', 'B', 'D'], 'Metabolite': ['C']}
diffusion_scores = run_diffusion({'Genes': ['A', 'B', 'D'], 'Metabolites': ['C']}, network)
3. You can also choose to provide a dataset with a column ‘Node’ containing node IDs as well as a column ‘logFC’ with their logFC. You may also add a ‘NodeType’ column to run diffusion by entity type.
Node |
LogFC |
|---|---|
A |
4 |
B |
-1 |
C |
1.5 |
D |
3 |
Also as a dictionary of node:score_value :
{'A':-1, 'B':-1, 'C':1.5, 'D':4}
diffusion_scores = run_diffusion({'A':-1, 'B':-1, 'C':1.5, 'D':4})
Combining point 2., you can also indicating the node type:
Node |
LogFC |
NodeType |
|---|---|---|
A |
4 |
Gene |
B |
-1 |
Gene |
C |
1.5 |
Metabolite |
D |
3 |
Gene |
Also as a dictionary of type:node:score_value :
{Gene: {A:-1, B:-1, D:4}, Metabolite: {C:1.5}}
diffusion_scores = run_diffusion({Gene: {A:-1, B:-1, D:4}, Metabolite: {C:1.5}}, network)
4. Finally, you can provide a dataset with a column ‘Node’ containing node IDs, a column ‘logFC’ with their logFC and a column ‘p-value’ with adjusted p-values. You may also add a ‘NodeType’ column to run diffusion by entity type.
Node |
LogFC |
p-value |
|---|---|---|
A |
4 |
0.03 |
B |
-1 |
0.05 |
C |
1.5 |
0.001 |
D |
3 |
0.07 |
This only accepted pythonicaly in dataaframe format.
See the sample datasets directory for example files.
Custom-network example¶
Source |
Target |
Relation |
|---|---|---|
A |
B |
Increase |
B |
C |
Association |
A |
D |
Association |
You can also take a look at our sample networks folder for some examples.
Input Mapping/Coverage¶
Even though it is not relevant for the input user usage, taking into account the input mapped entities over the background network is relevant for the diffusion process assessment, since the coverage of the input implies the actual entities-scores that are being diffused. In other words, only the entities whose labels match an entity in the network will be further processed for diffusion.
Running diffusion will report the mapping as follows:
Mapping descriptive statistics
wikipathways:
gene_nodes (474 mapped entities, 15.38% input coverage)
mirna_nodes (2 mapped entities, 4.65% input coverage)
metabolite_nodes (12 mapped entities, 75.0% input coverage)
bp_nodes (1 mapped entities, 0.45% input coverage)
total (489 mapped entities, 14.54% input coverage)
kegg:
gene_nodes (1041 mapped entities, 33.80% input coverage)
mirna_nodes (3 mapped entities, 6.98% input coverage)
metabolite_nodes (6 mapped entities, 0.375% input coverage)
bp_nodes (12 mapped entities, 5.36% input coverage)
total (1062 mapped entities, 31.58% input coverage)
reactome:
gene_nodes (709 mapped entities, 23.02% input coverage)
mirna_nodes (1 mapped entities, 2.33% input coverage)
metabolite_nodes (6 mapped entities, 37.5% input coverage)
total (716 mapped entities, 22.8% input coverage)
total:
gene_nodes (1461 mapped entities, 43.44% input coverage)
mirna_nodes (4 mapped entities, 0.12% input coverage)
metabolite_nodes (13 mapped entities, 0.38% input coverage)
bp_nodes (13 mapped entities, 0.39% input coverage)
total (1491 mapped entities, 44.34% input coverage)
To graphically see the mapping coverage, you can also plot a heatmap view of the mapping (see views). To see how the mapping is performed over an input pipeline preprocessing, take a look at this Jupyter Notebook or see process_input docs in DiffuPy.
Output format¶
The returned format is a custom Matrix type, with node labels as rows and a column with the diffusion score, which can be exported into the following formats:
diffusion_scores.to_dict()
diffusion_scores.as_pd_dataframe()
diffusion_scores.as_csv()
diffusion_scores.to_nx_graph()